FAQs about SerpentinaDB
What software do I need to take full advantage of SerpentinaDB?
Almost all features of the SerpentinaDB web site require a modern web browser with JavaScript and cookies enabled.
Here is a list of tested browsers grouped by desktop operating systems:
- Mozilla Firefox 4+
- Chrome 10+
- Internet Explorer 7+
- Safari 3+
What were the different languages used to develop SerpentinaDB?
- Design: HTML and CSS
- Dynamic Allocation: jQuery, JavaScript, and AJAX
- Database: MySQL
- Query connectivity: PHP
What softwares are required for perfect functioning of Jmol?
All browsers were installed with the latest JAVA plugins. In case of any discrepancy, please install latest JAVA plugin.
I get the following error message for Jmol visualisation → "The JAVA applet has been blocked because it is an untrusted application". What should I do?
This problem might arise in certain browsers due to the exceptions maintained by respective browsers.
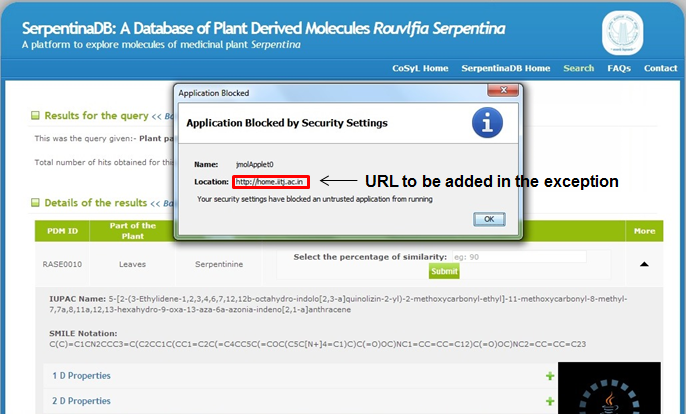
In such case please follow the following instructions:
Windows OS:
- Click on START
- Type "Configure JAVA"
- Click the "SECURITY" tab
- Click "Edit site list" at the bottom
- A window will open having an option "Add". Click on "Add"
- Another window will open. After the red exclamatory mark please type the entire URL displayed in the error. (In the image above the error is from http://home.iitj.ac.in)
This will clear the exception unblocking JAVA applets displayed from a particular website. But when you run the webpage for the first time you may get following error:
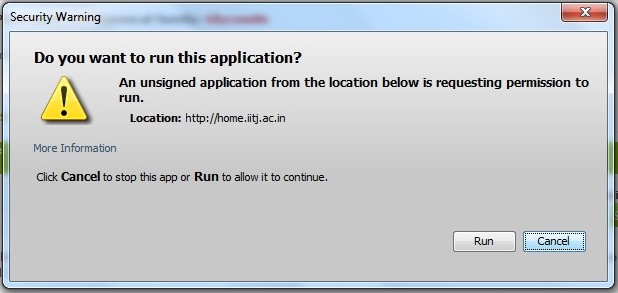
Click 'Run'. After this the visualization should work perfectly.
What is the source of the Cascading Style Sheet (CSS) template used for SerpentinaDB?
Click here to be directed to the page containing the CSS template.
What are different Plant Parts reported in SerpentinaDB?
Following plant part categories were identified based on reports:
- Stem
- Leaves
- Roots
- Root Bark
- Hybrid Cell Culture
- Cell Suspension Culture
- Root Culture
- Cell Culture
- Undried Roots
- Dried Roots
- Hairy Root Culture
- Unspecified
The PDM entry was classified as 'Unspecified', when no specific plant part, from which it was extracted, was reported.
How were the R. serpentina PDMs compiled?
In order to build an extensive library of PDMs from R. serpentina, data were compiled from literature and following web resources:
A database on antidiabetic plants, Global Information Hub On Integrated Medicine and India Herbs.
PubMed (http://www.ncbi.nlm.nih.gov/pubmed) was searched with the keywords 'Rauvolfia serpentina' and 'Rauwolfia serpentina' to obtain relevant literature.
Additionally, PhD dissertations reporting alkaloid constituents of R. serpentina, and following books were used for curation of data:
'The Alkaloids', 'The Alkaloids: Chemistry and Physiology' and 'The Alkaloids: Chemistry and Physiology'.
Two variants of spelling were used to address degeneracy in the literature.
All the resources were manually curated to extract data of PDMs and their additional details including chemical name, plant part, IUPAC
(International Union of Pure and Applied Chemistry), and 2D structure. The chemical structures of molecules were drawn and edited using MarvinSketch v5.10.0 software (https://www.chemaxon.com), and were subjected to energy minimization with Merck Molecular Force Field (MMFF94) using OpenBabel 2.3.1 software. To authenticate the chemical details obtained, molecules were also ascertained from the Dictionary of Natural Products (DNP).
To remove the redundant entries, data from all the resources were merged and an extensive library of 147 PDMs was compiled.
A separate entry was created for molecules that were obtained from more than one plant part leading to 227 individual entries.
What is Jmol?
Jmol is an open-source Java viewer for chemical structures in 3D, that does not require 3D acceleration plugins. Jmol returns a 3D representation of a molecule that may be used as a teaching tool, or for research e.g. in chemistry and biochemistry. It is free and open source software, written in Java and so it runs on Windows, Mac OS X, Linux and Unix systems.
Few technical advantages of Jmol are:
- Molecular 3D visualisation
- Zooming facilities
- Provision to download the viewed molecule in mol2 format
For furthur information please visit the Jmol website: Here
How to I download mol2 files for R. serpentina PDMs?
An interactive Jmol console is installed to enable 3D visualisation of the molecule. Right click on the Jmol window → File → Save copy of mol2.
What is the ZINC Similarity search?
ZINC is a free public resource for discovery of drug-like molecules.
The database contains over twenty million commercially available molecules in biologically relevant representations that may be downloaded in popular ready-to-dock formats and subsets. The similarity search done against ZINC compares the input SMILE and each SMILE present in the database.
Depending upon the similarity percentage (Eg: 90% which is the default threshold) it displays the entries for similar compounds.
For further information please visit the ZINC database: Here
What are 'ADMET properties', displayed as user output?
All ADMET properties were extracted using the Discovery Studio (DS) application.
The following properties were obtained using DS:
- Human intestinal absorption
- Aqueous solubility
- ADMET_Solubility
- ADMET_Solubility_Level
- Blood brain barrier penetration
- ADMET_BBB
- ADMET_BBB_Level
- Plasma protein binding
- ADMET_EXT_PPB
- ADMET_EXT_PPB#Prediction
- ADMET_EXT_PPB_Applicability
- ADMET_EXT_PPB_Applicability#MD
- ADMET_EXT_PPB_Applicability#MDpvalue
- CYP2D6 binding
- ADMET_AlogP98
- ADMET_PSA_2D
- ADMET_Unknown_AlogP98
- Hepatotoxicity
- ADMET_EXT_Hepatotoxic
- ADMET_EXT_Hepatotoxic#Prediction
- ADMET_EXT_Hepatotoxic_Applicability
- ADMET_EXT_Hepatotoxic_Applicability#MD
- ADMET_EXT_Hepatotoxic_Applicability#MDpvalue
For furthur information please visit the Accelrys Discovery Studio 4.0: Here
How to download the entire repository of R. serpentina PDMs?
Click here to download the entire repository of mol2 Files: Download
